Genomic scan for single nucleotide polymorphisms reveals patterns of divergence and gene flow between ecologically divergent species
- Stölting, Kai N. Department of Biology, Unit of Ecology & Evolution, University of Fribourg, Switzerland
- Nipper, Rick Floragenex, Portland, OR, USA
- Lindtke, Dorothea Department of Biology, Unit of Ecology & Evolution, University of Fribourg, Switzerland
- Caseys, Celine Department of Biology, Unit of Ecology & Evolution, University of Fribourg, Switzerland
- Waeber, Stephan Department of Biology, Unit of Ecology & Evolution, University of Fribourg, Switzerland
- Castiglione, Stefano Department of Chemistry, University of Salerno, Fisciano, Italy
- Lexer, Christian Department of Biology, Unit of Ecology & Evolution, University of Fribourg, Switzerland
-
15.01.2013
Published in:
- Molecular Ecology. - 2013, vol. 22, no. 3, p. 842–855
English
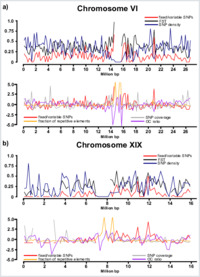
Recent advances in population genomics have triggered great interest in the genomic landscape of divergence in taxa with ‘porous’ species boundaries. One important obstable of previous studies of this topic was the low genomic coverage achieved. This issue can now be overcome by the use of ‘next generation’ or short-read DNA-sequencing approaches capable of assaying many thousands of single nucleotide polymorphisms (SNPs) in divergent species. We have scanned the ‘porous’ genomes of Populus alba and Populus tremula, two ecologically divergent hybridizing forest trees, using >38 000 SNPs assayed by restriction site associated DNA (RAD) sequencing. Windowed analyses indicate great variation in genetic divergence (e.g. the proportion of fixed SNPs) between species, and these results are unlikely to be strongly biased by genomic features of the Populus trichocarpa reference genome used for SNP calling. Divergence estimates were significantly autocorrelated (P < 0.01; Moran's I up to 0.6) along 11 of 19 chromosomes. Many of these autocorrelations involved low divergence blocks, thus suggesting that allele sharing was caused by recurrent gene flow rather than shared ancestral polymorphism. A conspicuous low divergence block of three megabases was detected on chromosome XIX, recently put forward as an incipient sex chromosome in Populus, and was largely congruent with introgression of mapped microsatellites in two natural hybrid zones (N > 400). Our results help explain the origin of the ‘genomic mosaic’ seen in these taxa with ‘porous’ genomes and suggest rampant introgression or extensive among-species conservation of an incipient plant sex chromosome. RAD sequencing holds great promise for detecting patterns of divergence and gene flow in highly divergent hybridizing species.
- Faculty
- Faculté des sciences et de médecine
- Department
- Département de Biologie
- Language
-
- English
- Classification
- Biological sciences
- License
- License undefined
- Identifiers
-
- RERO DOC 31841
- DOI 10.1111/mec.12011
- Persistent URL
- https://folia.unifr.ch/unifr/documents/302871
Other files
Statistics
Document views: 93
File downloads:
- lex_gss.pdf: 214
- FigS1-1_FigS2_TblS1_TblS2_TblS6.pdf: 147
- TableS3_hungary.txt: 45
- TableS3_italy.txt: 46
- TableS4.txt: 43
- TableS5.txt: 55